- IEEE Floating Point Standard
- Arrays
- Numerics
- Random Numbers
- Interpolation
- Integration
- Differentiation
- Scaling
- Linear Algebra
- Eigensystems
- Root-Finding
- Minimization
- FFTs
- ODEs
- PDEs
- MCMC (Markov Chain Monte Carlo)
IEEE Floating Point Standard
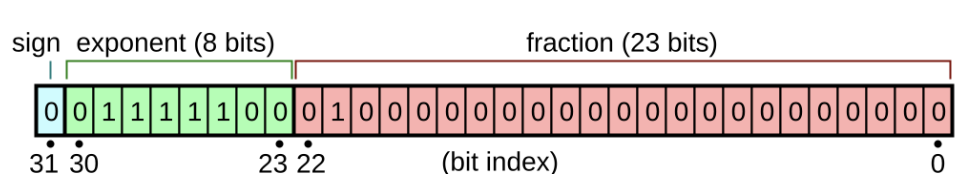
- $\pm 1+ 2^{e-127}\Sigma_{i=0}^{22} f_{i} 2^{-(i-23)}$
- e is the exponent number
- $f_{i}$ represents the floating bits
- Some edge cases:
- e = 0, $f \neq 0$ are called subnormal numbers, where “1” above is replaced with 0 and $e-127$ is replaced by $e-126$
- e=0 and f=0 is a signed zero
- e=255, f=0 is a signed $\inf$
- e=255, $f\neq = 0$ is NaN
Arrays
- TL;DR if you want to go fast, then use numpy arrays, since they are more contiguous in memory (and hence cache friendly) compared to python lists
- You also don’t have to worry about type checking since Python needs to check if operations
- There are also some built-in functions that numpy provides which are faster than Python built-ins
Numerics
- TL;DR Keep numbers from being too small and too large and you’re good
- $E[\delta (f(a))^{2}] \approx E[[\frac{\partial f}{\partial a}]^{2} \delta a^{2}]$
- Assuming similar errors for each operations, you find that the round-off error scales as $\sqrt{N}$
- Making finer time steps yields a better approximation, but the round-off error degrades this. There is some optimal step size which yields the most accurate result
Random Numbers
- Probability distributions have the defining property that $\int_{-\infty}^{\infty} dx P(x) = 1$
LCM (Linear Congruent Method)
- $x_{i+1} = (ax_{i}+c) mod M$
- a,c and M are parameters. After M random numbers, you repeat yourself
- a and M are choosen such that a*M is less than the highest integer available
- a,c and M are parameters. After M random numbers, you repeat yourself
- Numpy uses Mersenne Twister, which has a longer period (ie. number of cycles before generator repeats cycles back to the start)
Testing RNG
- The expectation value of the number of values to fall in each bin is $\frac{N}{n_{bins}} = \bar{N_{bin}}$
- With enough bins and enough data, each bin should follow a Poisson distribution. Hence, we expect the variance of each bin to be $\bar{N_{bin}}$
Nonuniform Distributions
- You can generate these from uniform ones. A generic procedure is as follows
- You have the uniform distribution $f(r) dr = dr$ for $0<r<1$ and you want to find $f(x) dx$
- For each little differential, $|f_{r} dr| = |f_{x} dx|$
- Rearranging yields: $|\frac{dx}{dr}| = \frac{1}{f(x)}$
- Solving the aoove for $x(r)$ allows one to transform the uniform distribution to the desired nonuniform one
- The above doesn’t always work. A notable example is gaussians. You get a non-invertible function. You do some strange transformations (Box-Muller method) to allow you to circumvent this
Non-Analytic Nonuniform Distributions
- You try and write down the cumulative distribution of your desired non-analytic distribution:
- $F(x) = \int x dx’ f(x’)$ which maps to the range $[0,1]$
- You can then generate points from f(x) by drawing from a uniform distribution and then using the formula $x = F^{-1}(r)$ where the inverse is calculated numerically via interpolating a tabulation of F(x)
Interpolation
- Interpolation means given some data, how do you generate estimates for data points that are not in the set
- In a linear framework, we want to find $\hat{f}(x) = \Sigma_{i} W(x, x_i) f(x_{i})$
Linear Interpolation
- Dead simple
- $\hat{f}(x) = f(x_{i}) + [f(x_{i+1}) \frac{x-x_{i}}{x_{i+1}-x_{i}}]$
- This corresponds to a weight of $W(x,x_{i}) = 1-|x-x_{i}|$
- Outside the range $-1 < (x-x_{i}) < 1$, w is 0
- Triangular function
Fitting Basis Coefficients
- Can think of $\hat{f}(x) = \Sigma_{i} a_{i} B_{i}(x_{j})$ where B_{i} is some set of basis functions with coefficient $a_{i}$
- Can think of this as a matrix equation: $\vec{f} = B\cdot \vec{a}$
- Solving yields: $\vec{a} = B^{-1} \cdot \vec{f}$
- You can invert B if the basis vectors are independent and have support where your samples are
- Hence $\hat{f}(x) = \Sigma_{i} B_{i}(x) \Sigma_{j} B_{ij}^{-1} f(x_{j})$
- $W(x,x_{i}) = B_{i}(x) \Sigma_{j} B_{ij}^{-1}$
- For linear interpolation, B is just the identity matrix
Polynomial fits
- Your basis functions could be other things. Take polynomials for instance:
- $B_{0} = 1$
- $B_{1} = x$
- $B_{2} = x^{2}$
- etc
- To help with numerical stability, you can rescale x to x’ by a constant so that powers of x’ are close to unity
- You can also rescale to the range -1 to 1
- Might as well take the polynomials to be the Legendre polynomials, which are defined on that interval
- For very high order, the matrix behaves poorly numerically
Fourier Basis
- The weights for the Fourier basis are $W(x,x_{i}) = \frac{1}{2\pi} \int_{-pi}^{\pi} d\omega exp[i\omega(x-x_{i})] = \frac{\sin[\pi(x-x_{i})]}{\pi(x-x_{i})}$
Integration
- The goal is to approximate $\int_{a}^{b} dx f(x) = \Sigma_{i=1}^{N} f(x_{i}) w_{i}$
Trapezoid Rule
- Each term in the sum takes the form $\frac{1}{2} \delta x (f_{i} + f_{i+1})$
Simpson’s Rule
- The main idea: we try to approximate the function locally around the points i-1, i, i+1 as a quadratic:
- $f(x) = \alpha’+\beta’ x + \gamma’ x^{2}$
- Make the transformation to $f(x) = \alpha + \beta (\frac{x-x_{i}}{dx})+ \gamma (\frac{x-x_{i}}{dx})^{2}$
- evaluate f at each point yields
- $f_{i-1} = \alpha -\beta +\gamma$
- $f_{i} = \alpha$
- $f_{i+1} = \alpha + \beta + \gamma$
- Inverting the above yields
- $\alpha = f_{i}$
- $\gamma = \frac{f_{i+1} + f_{i-1}}{2} - f_{i}$
- $\beta = \frac{f_{i+1} - f_{i-1}}{2}$
- You apply this approximation to two segments at a time, which require that N is odd
Rescaling Integrals
- You can rescale an integrals bounds:
- $I = \int_{a}^{b} dx f(x) \frac{b-a}{b’-a’} \int_{a’}^{b’} dx’ f(x(x’))$
- $x’ = (x-a) \frac{b’-a’}{b-a}+a'$
- Setting a’ to -1 and b’ to 1 is very useful. This yields
- $I = \frac{b-a}{2} \int_{-1}^{1} dx’ f(x(x’))$
- If you make the substitution: $x = q \frac{1+x’}{1-x’}$, then you can rescale an infinite range to a finite one:
- $I = \int_{0}^{\infty} dx f(x) = \int_{-1}^{1} dx’ \frac{2q}{(1-x’)^{2}} f(x(x’))$
- q is a choice to be made, and x=q when x’ =0
- You want to choose q somewhere near the halfway point in the integral
Differentiation
- $\frac{df}{dx} = \frac{f(x+\frac{dx}{2})-f(x-\frac{dx}{2})}{dx}$
- The central difference approximation of the derivative of f at x
- That’s it. There are other schemes that you can use (like forward difference and backwards difference), but those are more niche
- To extend to multivariable, you just fix the rest of the variables and only vary one at a time
- There is ostensibly an optimal choice for dx, which relates to the machine precision and the third derivative, but for all practical purposes, this is irrelavent since if you know the third derivative, why would you be approximating anything?
- There is something called jax, which lets you programatically calculate the derivative of a function using the chain rule
- The idea is that you calculate the analytical derivative of the function in parallel with the function itself. This process can be recursive, hence allowing the iteration
Scaling
- You can rescale all of the linearly independent units of a problem in order to accomplish two things
- Maximize the dynamic range by keeping numbers close to unity
- Allowing one to apply the same equations to different scales
- Take Poisson’s equation for gravity:
- $\nabla^{2} \Phi = 4\pi \rho G$
- We can redefine $r’ = \frac{r}{R}$
- $m’ = \frac{m}{M}$
- $t’ = \frac{t}{t_{0}} = \frac{t}{\sqrt{\frac{GM}{R^{3}}}}$
- The above let’s use rewrite Poisson’s equation in a manner which is scale invariant
Linear Algebra
LU
- Central problem: Solve Ax=b where A is a matrix and b is a vector
- Can use LU decomposition: A=LU where L is a lower triangular matrix and U is an upper triangular matrix
- L is a lower triangular matrix whose diagonal elements are 1 (this follows since there are $N^{2}+N$ values to set in L and U, but only $N^{2}$ contraints in A)
- This factorization is done by using Gaussian elimination with pivoting
- Once you have the LU factorization, you can easily find the determinant of A, since det(L) = 1 and det(U) = $\Pi_{i} U_{ii}$, and det(A) = det(L)det(U)
- Numerically, L and U are easier to work with than A (more 0s and 1s)
SVD
- If A is very close to being singular (causing inverses to be unstable), or Ax=b is not unique or doesn’t exist, you can use the SVD to get an approximate solution
- You can write any matrix as $A = U W V^{T}$
- U is an MxN matrix whose columns are orthonormal
- W is a NxN diagonal matrix
- $V^{T}$ is the transpose of an orthonormal matrix
- You can think of the SVD as telling you the range of A (ie. the subspace that A can map into) and the null space of A (ie. the space for which Ax = 0)
- The columns of U for the orthonormal basis of the range of A
- The columns of V correspond to the basis which spans the null space
- W tells you the rank of the matrix (the number of non-zero $w_{j}$)
- From the SVD, you can get the condition number of a matrix (ie. $C = \frac{max(W)}{min(W)}$)
- A singular matrix would have C be infinity. if 1/C approaches machine precision, then the matrix is singular enough that doing numerics on it directly is unstable
- You can easily get the inverse of A from the SVD: $A^{-1} = V W^{-1} U^{T}$
- $W^{-1}$ is trivial to find (take the inverse of all the diagonal elements)
Eigensystems
- Solve the equation $Ax = \lambda x$
- Technically, these are right eigenvectors. The left eigenvectors are defined as $xA = \lambda x$. These left ones have the same eigenvalues as the right
- Analytically, you just solve $det(A-\lambda I)=0$, but this is an awful way of doing this numerically. You actually use QR factorization. We don’t talk about this in class though
- The idea is that you find the QR decomposition, where Q is an orthogonal matrix (ie. $Q^{T} = Q^{-1}$) and R is an upper triangular matrix
- analytically, you use Gram-Schmidt, but this is numerically unstable. More numerically stable methods, like Givens rotations and Householder reflections, find ways to stuff zeros in the intermediate matrices to prevent round-off errors
- You then define the matrix B = RQ.
- Why? Observe that $B = RQ = Q^{-1}QRQ = QAQ = Q^{T}AQ$
- This means that B is a similar matrix to A, and hence has the same eigenvalues
- You apply this process iteratively, and since all the intermediate matrices are similar, they all have the same eigenvalues
- Note that this process doesn’t preserve the eigenvectors of A. You can find those by solving the characteristic equation ($Ax= \lambdax$) for the particular eigenvalue (potentially solving for multiple lambda at once with the block power method)
- Why? Observe that $B = RQ = Q^{-1}QRQ = QAQ = Q^{T}AQ$
- The idea is that you find the QR decomposition, where Q is an orthogonal matrix (ie. $Q^{T} = Q^{-1}$) and R is an upper triangular matrix
Uses
Principal Component Analysis (PCA)
- For a given real symmetric, positive definite matrix (this kind of thing shows up a lot in physics, like in the moment of inertia tensor), you can find the three principal axes of the system via eigendecomposition
Dimensional Reduction
- PCA is also useful for reducing the dimensionality of a dataset, which allows a lower dimensional approximation of a high dimensional dataset (for compression or interprability reasons)
- Suppose that you have some dataset $\vec{q_{i}}$ with mean $\vec{q_{0}}$
- You can define the residuals as $\vec{r-{i}} = \vec{q_{i}}- \vec{q_{0}}$ and the Covariance matrix as $C = \vec{r_{j}}\vec{r_{i}}^{T}$
- The eigenvalues and vectors tells you the coordinate system which diagonalizes this matrix
- Since eigenvectors with larger eigenvalues have large variance, they are more important to describing the shape of the data. Hence, you can throw away the smaller eigenvalues and associated eigenvectors, and still keep the majority of the information
Root-Finding
- Solve the equation $f(x) = 0$
- For many methods, you need to bracket the root (ie. define a region on the domain of x which has the root in it).
- Typically, you just start with an initial guess for a bracket, then grow the bracket symmetrically by some factor until you have the root. This doesn’t always work though
- The brain-dead way of finding a root is bisection. This is just binary search, with your stopping condition being if the new bracket’s width is on the order of machine precision
- There is a less-sure fire, but faster method called Brent’s method, which fits a parabola to the left, right and midpoint of your bracket, then steps towards the nearest root of the parabola
- If the root falls outside the bracket, or the update step is very tiny, then Brent defaults to bisection
- The exact details do some massaging of terms to make things numerically stable when solving the quadratic
- You you have access to the derivative of the function, you can use Newton-Raphson’s method
- You iteratively find $x_{i+1} = x_{i}-\frac{f(x)}{f’(x)}$
- If you are nowhere near a root, then Newton’s method can not converge, so it’s usually best to use for really precise determinations of a root, due to it’s quadratic convergence
Minimization
Quadratic Fit
- Assume that you can evaluate your function f(x), and you have defined your bracket from [a,c], and you choose some point b in the bracket
- Basic idea:
- Try fitting a quadratic of the form $\tilde{f}(x) = \alpha x^2 \beta x +\gamma$ to the points a,b,c (ie. solve for the parameters via solving a matrix equation).
- Define your new bracket according to the minimum of this quadratic.
- This isn’t perfectly stable, since there is a change that the minimum falls outside of your current bracket
Golden Section Search
- Another idea: We want to shrink the bracket by a consistent amount each step
- Define the fractional position of x=b as $w = \frac{b-a}{c-a}$
- Suppose that the the new trial x is greater than b. Define the fractional position of the trail x as $z = \frac{x-b}{c-a}$
- We want the new bracket length ($z+w$) to be the same as the original bracket length $1-w$
- If we apply this requirement iteratively, we want the fractional position of the new trail x to be the same as b was in the old bracket
- In math, this means $\frac{z}{1-w} = w$
- Solving for w, we see that $w = \frac{3-\sqrt{5}}{2}$
- A more standard 1D minimization routine combines Quadratic fitting and Golden Section Search
- If the parabolic step falls outside the bracket, or the step is larger than the previous step, you fall back on GSS
- this is called Brent’s method, confusingly enough…
Multi-Dimensional
- Idea: We use the Hessian matrix ($A_{ij} = \frac{\partial^{2} f}{\partial x_{i}\partial x_{j}}$) to approximate the function locally as a multi-dimensional quadratic, then step to the minimum
- Problem: You normally don’t know the Hessian of a function
- Idea: If we know the eigenvectors of the Hessian, then you can minimize along the direction of each eigenvector (see here)
- If you don’t know the Hessian, then you can use Quasi-Newtonian methods like Broyden-Fletcher-Goldfarb-Shanno
FFTs
- Definition of Fourier Transform:
- $H(f) = \int_{-\infty}^{\infty} dt h(t) exp(2\pi i ft)$
- $h(t) = \int_{-\infty}^{\infty} dt H(f) exp(-2\pi i ft)$
- Discretization:
- Suppose that your set of samples at equally spaced locations $t_{k} = k\Delta$, where k runs from 0 to N-1
- Let $f_{n} = \frac{n}{N\Delta}$, where n ranges from $-\frac{N}{2}$ to $\frac{N}{2}$
- $H(f_{n}) \approx \Delta \Sigma_{k=0}^{N-1} h(t_{k}) exp(\frac{2\pi i kn}{N})$
- $h_{k} = \frac{1}{N} \Sigma_{n=0}^{N-1} H_{n} exp(\frac{-2\pi i kn}{N})$
- This is slow to calculate (scales as $N^{2}$). FFT can give $N log N$ scaling instead
- Basic Idea: Assumes that N is a power of 2 ( can pad out dataset to make it a power of 2). Split the series into even and odd terms. Recursively apply this splitting of even and odd terms. That’s it (lot’s of bookkeeping, but that’s it)
ODEs
- an Nth order differential equation can be reduced to N 1st order differential equations by letting $\frac{dx}{dt} = y$ (and similar constructs can be used for higher derivatives)
- To calculate first order derivatives, you should use Runga-Kutta methods
- This is good enough for most applications
- If you care about energy conservation, you can use things like Leapfrog
- If you have multiple time scales in a system, then you need to make surer that your step-size is small enough to capture the relevant phenomena
PDEs
Initial Condition Problems
- Consider PDEs of the form $\frac{\partial \vec{q}}{\partial t} = -\frac{\partial \vec{F}(q)}{\partial x}$, where F is the conserved flux. For simplicity, let’s look at the example $\frac{\partial \vec{q}}{\partial t} = -v\frac{\partial q}{\partial x}$
- The naive way of solving this numerically is discretizing space and time into equal spaces:
- $x_{j} = x_{0}+j\delta x$
- $t_{j} = t_{0}+n\delta t$
- This gives an update scheme like: $\frac{q_{j}^{n+1}-q_{j}^{n}}{\delta t} = -v \frac{q_{j+1}^{n}-q_{j-1}^{n}}{2\delta x}$
- If we are given q defined for all j at n=0, then we can propagate outwards to solve for when n=1, n=2, etc.
- This is not stable. We can see this by Taylor expanding around the difference equation solution (which is $q_{j}^{n} = \epsilon^{n}exp(ikj\delta x)$)
- Solving for $\epsilon$, we see that the magnitude of $\epsilon$ is greater than 1 for all k, which implies instablity
- The Lax method simply replaces the estimate of $q_{j}$ at step n with the average of the neighboring points
- This is stable, as long as $\frac{\delta x}{\delta t} < v$, which is calculated by the same sort of stability analysis as before (called von Neumann Analysis)
- This is called the Courant condition. Physically, this means that the time step of the simulation must be smaller than the propagation speed of the signal across a single cell
- This has a problem, in that higher order wavenumbers get dampened out
- You can see this by rewriting the lax equation to tease out a term which looks like $\frac{\delta x^{2}}{2\delta t}\frac{\partial^{2}q}{\partial x^2}$ (like a viscosity term)
- Implicit methods also exist, where we utilize future mesh points in the difference equations in such a way that they can’t be uncoupled
- For instance, the diffusion equation ($\frac{\partial q}{\partial t} = \frac{\partial }{\partial x}(D \frac{\partial q}{\partial x})$) can be discretized like $q_{j}^{n+1} = q_{j}^{n}+\alpha(q_{j+1}^{n}-2q_{j}^{n}+q_{j-1}^{n})$
- These are just a linear system of equations with N unknowns (the $q_{j}^{n+1}$)
- Crank-Nicolson methods are a combination of the two (ie. you take a half step explicitly, and a half step implicitly). The overall method is still implicit, but you get better stability properties and conservation of small-scale structure
Boundary Condition Problems
- If you have constant coefficients, you can get away with Fourier methods
- Look at Poisson’s equation: $\nabla ^{2} q = \rho$
- The Fourier transform of $\nabla^{2} q = -k^{2} q$. Hence, we can write q as $q = -FT(\frac{\rho}{k^{2}})$
- After discretizing, we can apply a discrete fourier Transform to all the involved variables
- $q_{j} = \frac{1}{J}\Sigma_{n=0}^{J-1} \tilde{q}_{n}exp(\frac{-2\pi i j n}{J})$ and similar expansion for $\rho$
- Plugging in and rearranging, we can match coefficients to find that $\tilde{q_{n}} = \frac{\delta^{2}\tilde{\rho_{n}}}{2\cos(\frac{2\pi n}{J})-1}$. The original variables can be calculated via inverse Fourier Transforms
- Another category of methods are relaxation methods, where, if you are dealing with elliptic PDEs (ie. $Lq = p$ where L is an elliptic operator), you can instead solve the diffusion equation $\frac{\partial q}{\partial t} = Lq-p$.
- Static solutions to the above satisfy the original eqaution, so if you start with a random q, then iteratively apply the finite difference method of the diffusion equation until q doesn’t change anymore (you are “relaxing” to the stable solution)
MCMC (Markov Chain Monte Carlo)
- Suppose that you have some data $\vec{d}$ and a set of model parameters $\vec{theta}$. MCMC allows you to generate a distribution of $\theta$ (called the posterior) which is peaked at the “best” choice of parameters, whose spread is associated with uncertainties about the correct parameters
- This makes no assumption on wheather $\theta$ is properly modelling the data
- From Baye’s Theorem: $P(\vec{\theta}| \vec{d}) = \frac{P(\vec{d}|\vec{\theta})P(\vec{\theta})}{P(\vec{d})}$
- In words: The probability distribution of the real $\theta$ given the data (posterior) equals the probability distribution of the data given the model parameters (likelihood) multiplied by the distribution of the model parameters (prior) divided by the distribution of the data (marginalization)
- in fewer words: posterior = likelihood*prior/marginalization
- Intuitively, you can think of your current best guess for distribution of the model parameters (say, based on past data you have collected)
- The likelihood represents our current understanding of how the model parameters describe the data (ie. our model)
- The marginalization is the distribution of data sets over all possible choices of $\theta$
- You don’t normally need to calculate this, but if you had to, it’s just an integral over all $\theta$
- $P(\vec{d}) = \int d^{N} \vec{\theta} P(\vec{d}|\vec{\theta}) P(\theta)$
- You don’t normally need to calculate this, but if you had to, it’s just an integral over all $\theta$
Markov Chain
- A Markov Chain is a sequence of points $\vec{\theta_{n}}$ such that the distribution of point n+1 only depends on point n
- We want this sequence to have the property that it is distributed according to the posterior
- For any desired distribution $\pi(\vec{\theta})$, we can get a Markov chain that follows $\pi$ if $\pi(\vec{\theta_{1}})p(\theta_{1}|\theta_{2}) = \pi(\vec{\theta_{2}})p(\theta_{2}|\theta_{1})$
- Proof: Ask the question: Given a set of points $\theta_{1}$, what is the distribution of the next set of points $\theta_{2}?$
- $p(\theta_{2}) = \int d^{N} \vec{\theta_{1}} p(\theta_{2}|\theta_{1}) \pi(\theta_{1})$
- Can use Markov chain property to show that $p(\theta_{2}) = \pi(\theta_{2})$
- So if the Markov property holds, then $\pi$ is an equilibrium distribution
Metropolis Hastings
- Algorithm used to generate a Markov Chain
- For each step, we generate some $\theta_{i}$, at which we evaluate the posterior $\pi(\theta_{i})$
- We generate a candidate $\theta_{i+1}$ from some proposal distribution $q(\theta_{i+1}|\theta_{i})$ (Gaussian is a common choice)
- We calculate the acceptance probability $\alpha(\theta_{i+1}|\theta_{i}) = min(1, \frac{\pi(\theta_{i+1})q(\theta_{i}|\theta_{i+1})}{\pi(\theta_{i})q(\theta_{i+1}|\theta_{i})})$. With randomly accept the candidate based on this probability
Burn-in and Proposal Distribution
- Your starting point in MCMC biases what your final distribution looks like
- You also need a finite number of steps before the equilibrium is reached
- So you need to figure out how many steps you need, and how may of these steps are independent. Typical tools for this are the autocorrelation function and the autocorrelation time
- You choice of proposal distribution also affects the burn-in time/convergence of the system
- a proposal distro which generates too large of steps might not converge
- a distro which generates small steps might take forever to converge